Abstract
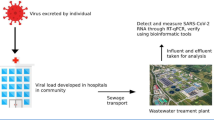
The COVID-19 pandemic has been monitored by applying different strategies, including SARS-CoV-2 detection with clinical testing or through wastewater-based epidemiology (WBE). We used the latter approach to follow SARS-CoV-2 dispersion in Tapachula city, located in Mexico’s tropical southern border region. Tapachula is a dynamic entry point for people seeking asylum in Mexico or traveling to the USA. Clinical testing facilities for SARS-CoV-2 monitoring are limited in the city. A total of eighty water samples were collected from urban and suburban rivers and sewage and a wastewater treatment plant over 4 months in Tapachula. We concentrated viral particles with a PEG-8000-based method, performed RNA extraction, and detected SARS-CoV-2 particles through RT-PCR. We considered the pepper mild mottle virus as a fecal water pollution biomarker and analytical control. SARS-CoV-2 viral loads (N1 and N2 markers) were quantified and correlated with official regional statistics of COVID-19 bed occupancy and confirmed cases (r > 91%). Our results concluded that WBE proved a valuable tool for tracing and tracking the COVID-19 pandemic in tropical countries with similar water temperatures (21–29 °C). Monitoring SARS-CoV-2 through urban and suburban river water sampling would be helpful in places lacking a wastewater treatment plant or water bodies with sewage discharges.
Similar content being viewed by others
Introduction
The mitigation and control of the current COVID-19 pandemic caused by the coronavirus SARS-CoV-2 have been challenging. Strategies to increase testing capabilities are urgent to better monitor disease transmission and plan health interventions accordingly. In some regions of the world, infrastructure, trained personnel, and reagents for clinical testing are limited. Wastewater-based epidemiology (WBE) is a valuable tool to follow community spread and monitor outbreaks and the results of public health interventions (Kitajima et al., 2020; Xagoraraki & O’Brien, 2020). WBE takes advantage of the genetic material of pathogens, or the pathogens themselves, being shed in the feces and urine of infected patients, which can be detected using molecular techniques such as qPCR and DNA sequencing (Berchenko et al., 2017; Farkas et al., 2018; Gulino et al., 2020; Lago et al., 2003). Recent studies reported the presence of SARS-CoV-2 RNA in human feces in patients with and without gastrointestinal symptoms (Gu et al., 2020; Kitajima et al., 2020; Song et al., 2020; Xiao et al., 2020). Specifically, several studies have detected the genomic region coding for the nucleocapsid (N) protein in SARS-CoV-2 WBE (Medema et al., 2020; Nemudryi et al., 2020; Wu et al., 2020). SARS-CoV-2 RNA concentrations in feces could be up to 108 copies per gram of feces (Lescure et al., 2020; Pan et al., 2020; Wölfel et al., 2020). As a result, SARS-CoV-2 has been detected in wastewater worldwide, e.g., Netherlands, France, Spain, USA, Australia, and India (Ahmed et al., 2020a, 2020b; Kumar et al., 2020; Medema et al., 2020; Randazzo et al., 2020; Wu et al., 2020; Wurtzer et al., 2020), with concentrations of up to 103 copies per mL. Experiments measuring SARS-CoV-2 decay in wastewater-detected RNA signals for up to 26.2 days (Ahmed et al., 2020a, 2020b; Bivins et al., 2020). Although it is still challenging to link the number of detected copies with the number of COVID-19 cases, the sensitivity of SARS-CoV-2 detection in wastewater is high, reaching 0.02–0.1% infected population or between 2 virus shedders per 10,000 persons and one virus shedder per 1000 persons (Jørgensen et al., 2020; Michael-Kordatou et al., 2020). Most research on SARS-CoV-2 in wastewater has focused on wastewater treatment plants (WTP) samples or rivers receiving treated water discharges. However, many human settlements in the developing world lack this kind of infrastructure, or such facilities are not efficient, resulting in raw wastewater being spilled directly into rivers or oceans (Aslan et al., 2020; Guerrero-Latorre et al., 2020; Rimoldi et al., 2020; Street et al., 2020). SARS-CoV-2 RNA has also been detected in sewage and WTP sludge (Kocamemi et al., 2020; Peccia et al., 2020). RNA concentration in primary sludge follows the changes in COVID-19 cases in the community, suggesting that sludge can also be used for SARS-CoV-2 WBE (Peccia et al., 2020).
Ideally, testing for SARS-CoV-2 should be carried out on each suspected infected individual. Still, due to a shortage of reagents or clinical testing capabilities, this is not possible in some areas of the world. Such an issue is present in Tapachula, a city of 217,550 habitants (INEGI, 2020) in southeast Mexico, in Chiapas State, at the border with Guatemala, which is a dynamic entry point for people from Central America and the Caribbean seeking asylum in Mexico or traveling to the USA. Facilities for SARS-CoV-2 testing are limited and expensive in this small border city, especially with the PCR method.
Migration flow is challenging to estimate in Tapachula. In 2020, Secretaría de Gobernación (Secretariat of the Interior or SEGOB, Mexico) reported that around 905,000 people entered Chiapas State, 857,000 of whom had been visiting Tapachula in 2020. However, these official data sub-estimate people crossing the city of Tapachula. The Comisión Mexicana de Ayuda a Refugiados (COMAR, 2021) reported more than 26,000 refugee applications in Tapachula in 2020, and more than 63,000 in 2021 (until September). Since these numbers do not include migrants who do not cross the border at the official entry point, the number of migrants is likely higher. Considering that, until January 2021, there were 1321 COVID-19 cases in Tapachula, it seems the actual number of cases is being sub-estimated. WBE could be a practical approach for estimating SARS-CoV-2 dissemination in cities or countries in similar conditions.
In this study, we explored the possibility of applying the principle of WBE to monitor the presence of SARS-CoV-2 RNA in sewage and urban rivers to estimate its presence and spread in the community. Mexico treats only 48% of its wastewater (CONAGUA, 2018). Even if sewage facilities in several places are available, sewage is not necessarily directed to a WTP. In Tapachula, wastewater treatment facilities operate at 37% capacity, effectively serving a small fraction of the population (INEGI, 2016). Although ideally, wastewater treatment should be improved in the region, in the meantime, it is necessary to investigate what type of water samples could be used for WBE. We analyzed water samples taken in urban rivers, sewage, WTP, and suburban rivers. This approach would help produce informative and valuable data on water contamination and validate a helpful method for other cities with similar settings (Pandey et al., 2021). Previous studies showed that SARS-CoV-2 tends to degrade faster in warmer wastewater temperatures, i.e., 25–37 °C (Ahmed et al., 2020a, 2020b), close to those registered in tropical environments like Tapachula, with a mean annual temperature of 28.5 °C (CONAGUA, 2020). Thus, it is crucial to establish methodologies to detect SARS-CoV-2 under tropical conditions because viral particles’ persistence might be shorter than those measured in northern latitudes.
Materials and Methods
Study Area
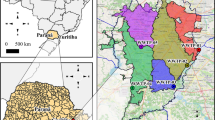
We carried out this study in the hydrographic basins of the Coatán and Cahoacán rivers, located in the Soconusco region in southeastern Mexico. The rivers of both basins originate in the Sierra Madre de Chiapas mountains. The Coatán basin belongs to two countries, 30 km is in Guatemala and 80 km on the Mexican side. The Cahoacán has its origin in the geography of the Tacaná volcano. Tapachula is a municipality located between these two river basins. The urban area has more than 200,000 inhabitants. Two tributaries cross it: the Coatancito river flows into the Coatán river, and the Texcuyuapan river that flows into the Cahoacán river. These two tributaries receive urban domestic raw sewage (Fig. 1).
Map showing sampling sites in Tapachula city. R1-R6 denotes rivers samples. PA1E and PA1S represent samples from the WTP at the wastewater entry and exit point, respectively. Circle sizes represent the average N1 copy number/mL detected per site along the sampling period. Site names are shown in Table 1
Sampling Sites
Hospital Discharges
We collected samples from sewage drains 40 m away from the local COVID-19 Hospital wastewater discharge (Davó et al., 2021), designated as H1 (N 14° 53′ 32.23″ W 92° 15′ 57.22″). Due to the limited communication with local health authorities, sampling in hospitals was restricted to four samples in the sewage outside the COVID-19 hospital.
Rivers Within the City
Samples were collected at two sites: Texcuyuapan river (R2), in the southeastern part of the city (N 14° 52′ 12.61″ and W 92° 15′ 44.90″), and Coatancito river (R4), in the southwestern part of the city (N 14° 54′ 7.67″ and W 92° 16′ 26.97″). Texcuyuapan river receives large amounts of domestic wastewater from surrounding areas in the city. After crossing the entire city, the Coatancito river is also considered a recipient of domestic sewage.
Rivers Outside the City
Four sampling sites were established outside the city: two for the Cahoacán basin (R1 and R3) and two for the Coatán (R5 and R6). Site R1 is approximately 25 km (straight line) northeast of Tapachula (N 15° 2′ 20.11″ and W 92° 10′ 13.92″). This site was chosen because it is expected to have a low anthropogenic impact. The R3 site (N 14° 49′ 31.06″ and W 92° 15′ 32.35″) is downstream in the city’s eastern part, approximately 8 km straight from the urban area. Most urban wastewater has been discharged into the basin, including treated water from COAPATAP-WTP (a municipal WTP). After this site, the river water is used to irrigate many crop sites in the region.
Site R5 was located approximately 10 km downstream of the city on the west side (N 14° 53′ 56.45″ and W 92° 21′ 1.64″), where Coatán river has collected all the water from the eastern part of the city. Site R6 (N 14° 51′ 57.03″ and W 92° 25′ 26.23″) was chosen because water downstream this point is used for several purposes along the river’s course, including human consumption. It provides water to the municipality of Mazatán, an urban settlement with a population of more than 20,000 people.
Municipality WTP (COAPATAP-WTP)
We collected water samples from the COAPATAP-WTP at two points: raw wastewater (PA1E) and exiting treated wastewater (PA1S).
Water Sampling
A total of 80 water samples were collected from September 2020 to January 2021 into sterile containers wearing personal protective equipment. We collected samples for RNA analysis in 250 mL polypropylene bottles with a screw cap. At the same time, we collected samples for TOC (total organic carbon) detection in 250 mL amber glass bottles with a Bakelite screw cap. Details about the number and type of samples taken are in Table 1. We used a bucket for sampling in the Coatán river due to poor accessibility. Before collection, the bucket previously sanitized was rinsed three times with water from the same river according to the guidelines of the Mexican regulation NMX-AA-003-1980 (SECOFI, 1980). Thanks to better accessibility in the Coatancito, Texcuyuapan, and Cahoacán rivers, samples were collected in counter-flow directly into the propylene and glass bottles.
At the COAPATAP-WTP, samples were collected in the influent (inlet) and the effluent (outlet), in both cases counter-flow. For collecting wastewater samples from the sewage of the COVID-19 Hospital (H1), the sewer’s cover was removed, and samples were collected with 1-L plastic containers previously sanitized and rinsed as above, obtaining the water from a pipe connected to the hospital drainage. All samples were kept at 4 °C for a maximum of 1 h during transportation to the laboratory. Glass bottles and polypropylene jars were stored at − 20 °C, and screw cap centrifuge tubes at 4 °C until further processing.
Physicochemical Determinations
We measured some parameters in situ electrical conductivity (EC, ms/cm) and temperature (°C) with a Horiba water checker U-10 multiparametric potentiometer; pH with a portable analytical instrument (EC-983 potentiometer); and dissolved oxygen (DO) using a Hach HQ30d Flexi electrode. We placed electrodes directly in the stream of the river or the WTP (inlet and outlet). Samples collected in the Coatán river and COVID-19 hospital drainage were measured by submerging the electrodes in the containers.
At the laboratory, total carbon (TC), total organic carbon (TOC), and inorganic carbon (IC) were detected by the total organic carbon analyzer Shimadzu TOC-VCSH.
Viral Concentration and RNA Extraction
We modified methods proposed by Ahmed et al. (2020a, 2020b) and Wu et al. (2020) for virus concentration. A 160 mL sample of water was homogenized by inversion. Then, we added 25 mL of glycine buffer (0.05 M, pH 9.0) plus meat extract (3% w/v), and we stirred it at 200 rpm for 30 min at 25 °C. The meat extract detaches viral particles from organic matter. The mixture was then centrifuged at 3200×g for 40 min at 4 °C to remove larger suspended solids. The supernatant was divided into four tubes with 40 mL each, and PEG 8,000 (10% w/v) + NaCl (2% w/v) were added to each of them. They were incubated for 2 h at 4 °C with constant stirring (150 rpm), then samples were centrifuged at 7740×g at 4 °C/1 h to precipitate the viral particles. The supernatant was discarded, and the pellet was resuspended in 250 µL of TRIzol® Reagent (Invitrogen; California, USA) to obtain a total of 1 mL of concentrate from the four tubes. Total RNA was extracted with a Purelink RNA Minikit (Ambion-by Life Technologies. California, USA) from the 1 mL TRIzol® concentrate, following the manufacturer’s instructions. RNA was eluted in 30 µL of nuclease-free water and stored at − 20 °C until later use.
Reverse Transcription and End-point PCR for Process Control
We considered RNA pepper mild mottle virus (PMMoV) detection as a positive control to confirm RNA extraction and amplification. This single chain RNA virus affects pepper plants and has been recognized as an indicator of water contamination by human feces (Kitajima et al., 2018); thus, it could be used as a marker of fecal contamination of water. cDNA was synthesized from 2 µL of the total RNA extracted using the Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Scientific, Massachusetts, USA). The final reaction volume was 10 µL. The reaction protocol for RT was 25 °C for 10 min, 50 °C for 30 min, and 85 °C for 5 min. The synthesized cDNA was used in PCR to amplify a 632 bp viral fragment from PMMoV using primers PMMoV Fw 5′-GGAAAACGCCTACACAGATCG-3′ and PMMoV Rv 5′-GTACGCACAATTGTTCAACG-3′ (Aguado-García et al., 2020). The 12 µL reaction included 0.2 pM of each primer, PCR Master Mix 1x (Promega), and 1 µL of cDNA. PCR protocol included denaturation at 94 °C for 5 min, plus 35 cycles of denaturation (94 °C 30 s), annealing (56 °C for 1 min), and extension (72 °C for 1 min), followed by a final extension step of 72 °C for 10 min. PCR products were visualized by electrophoresis in 1% agarose gels.
We used the PMMoV (final point PCR) and RNAse P fragments (qPCR, see below) as positive controls for PCR amplification. If we obtained a positive PCR result for any of these markers, we considered it inhibitor-free.
Quantitative PCR (qPCR)
We quantified the viral RNA in water samples with the set of primers and probes for the SARS-CoV-2N gene (N1 and N2) and the RNAse P marker present in all human samples, as designed by the CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel (IDT; IA USA) (CDC, 2020). One-step reactions were carried out with Luna® Universal One-Step RT-qPCR Kit (New England Biolabs, Massachusetts, USA) in a final volume of 10 µL, of which 2 µL correspond to the total RNA extracted from each sample. Each sample was assayed in triplicate for the three markers (N1, N2, and P gene) and included triplicate controls without RNA (NTC) in each run. All reactions were run in a CFX96™ Touch Real-Time PCR Detection System (Bio-Rad, CA, USA). Similar to other studies on environmental samples (Medema et al., 2020; Philo et al., 2021; Westhaus et al., 2021; Zhou et al., 2021), and as suggested by CDC (2020), samples with Cq less than 40 were taken as positive. We calculated the total viral load by interpolating the Cq value obtained in calibration curves from solutions of synthetic standards (plasmid controls) for N1, N2 (2019-nCoV Positive Control: nCoVPC), and RNase P (Hs_RPP30 Positive Control) (IDT; IA, USA) markers; with a range of 100,000 to 1.28 copies/µL (R2 > 0.99 for the three markers). Results were expressed as gene copy number per mL (copy number/mL) of water according to the standard curve for each marker.
Sanger Sequencing of qPCR Products
We selected samples with the highest copy number of N1 and N2 markers for cloning. qPCR products were visualized on 1.5% agarose gels; bands were excised and purified with Zymoclean™ Gel DNA Recovery Kit (Zymo Research. CA, USA). They were eluted in 15 µL of nuclease-free water, ligated to the pJET1.2 vector following the manufacturer’s instructions (CloneJET PCR Cloning Kit, Thermo Scientific, Massachusetts, USA) and transformed into calcium-magnesium competent cells of Escherichia coli DH5-α (F–, endA1, glnV44, thi-1, recA1, relA1, gyrA96, deoR, nupG, purB20, φ80dlacZΔM15, Δ(lacZYA-argF)U169, hsdR17 (rK– mK+), λ–), according to the protocol reported by Serrano-Rivero et al. (2013). Clones obtained were analyzed by colony PCR using vector primers: pJET-Fw (5′-CGACTCACTATAGGGAGAGCGGC-3′) and pJET-Rv (5′-AAGAACATCGATTTTCCATGGCAG-3′), with the following protocol: initial denaturation at 95 °C, for 3 min; followed by 94 °C, for 30 s; 60 °C, for 30 s; 72 °C for 1 min; for 30 cycles. Positive PCR clones were selected for plasmid extraction using the QIAprep® Miniprep Handbook kit (Qiagen. Maryland, USA) and sent for Sanger sequencing to the Synthesis and Sequencing Unit of the Institute of Biotechnology (La Unidad de Síntesis y Secuenciación de DNA (USSDNA), UNAM, Mexico).
Statistical Analysis
Because data did not follow a normal distribution, statistics were assayed with non-parametric tests. Correlation evaluations were analyzed with the Spearman-Rank test, presenting the correlation matrices with a visual method. All analysis and graphs were done with R software (R Core Team, 2020).
Results
Physicochemical Characteristics of the Water
We determined physicochemical parameters to verify the level of impact due to contamination in all water samples. 80 samples were collected and analyzed for water quality parameters and RNA extraction in nine sites (Fig. 1). The results reflect the anthropogenic impact on water bodies (Table 1). Organic matter concentration (measured as TOC) and dissolved oxygen were used as indicators of water pollution. The higher TOC and the lower dissolved oxygen indicate a higher pollution level. As expected, sewage outside COVID-19 Hospital (H1) and COAPATAP-WTP (PA1E) had the most polluted water samples. The least anthropogenic contaminated water was found upstream Cahoacán river (R1), with higher dissolved oxygen and lower TOC.
All sites showed neutral pH. Due to climatic conditions in the zone, the temperature was lower at Cahoacán river (R1, 21 °C). The highest temperature was measured in the water treatment plant (COAPATAP-WTP, PA1E), probably due to biological activity. The temperature in the effluent (PA1S, 29 °C) can be influenced by direct sunlight exposition through the chlorination tank.
SARS-CoV-2 RNA Detection and Quantification
PCR amplification of the PMMoV virus allowed us to verify the presence of viral genetic material in obtained extracts. We detected PMMoV in all samples, except samples taken in R1 and some samples obtained from R5 (80% positivity, Fig. 2). One sample from R1 did not amplify either PMMoV, SARS-CoV-2, or RNAse P, opening the possibility that it could contain PCR inhibitors. To test if this was the case or if there was not enough fecal contamination to be detected by the end-point PCR, we cleaned up RNA from negative qPCR samples through OneStep™ PCR Inhibitor Removal Kit (Zymo Research, USA) and repeated qPCR. PMMoV remained undetected, but we observed qPCR detection for RNAse P marker.
Positivity rate of SARS-CoV-2 (N1 and N2) and PMMoV RNA detection by RT-qPCR and end-point PCR, respectively, in the study sites during the entire sampling period, considering a Cq > 40 as a positive. Bar width correlates to the number of samples, while red and green correspond to negative and positive proportions, respectively
RT-qPCR tested the presence of SARS-CoV-2. It was detected in all sites using both genomic regions (N1 and N2, Table 1, Fig. 2), with 36.7% of samples producing positive results for both N regions. N1 showed a higher positivity rate and copy number/mL, with a positivity rate ranging from 25 to 100% and copy number/mL from < 2 up to 657. N2 positivity ranged between 9 and 88%, and copy number/mL obtained was < 2 up to 738. RNAse P, human contamination control was positive in more than 80% of samples analyzed (Fig. 2).
qPCR products amplified for the N1 and N2 regions were sequenced in four samples, collected in PAE1, R2, R4, and H1. The resulting sequences were aligned with the BLAST algorithm to the SARS-CoV-2 reference genome (NCBI accession number NC_045512.2). We obtained 100% similarities for N1, whereas N2 identity with the reference genome ranged from 97.1 to 100% along the corresponding region (Table S1).
Comparison of Active Cases and Detected Copy Number
Viral detection using the N1 region during the 12 weeks sampling period in wastewater treatment plant (influent, PA1E) and Texcuyuapan river (R2) showed a similar trend matching the estimated COVID-19 active cases, calculated as a moving 14-day average of daily new cases reported by the Ministry of Health (Secretaría de Salud, Mexico (https://datos.covid-19.conacyt.mx/#DownZCSV) (Fig. 3). PA1E shows the highest copy number in January 2021, reflecting the surge in COVID-19 estimated active cases in Tapachula (red line Fig. 3). However, the highest copy number value for N1 in PA1E was registered before the peak in estimated COVID-19 cases. On the other hand, samples from R2 show a slight delay in reaching the highest values during the sampling period.
Comparison of N1 copy number per mililiter with estimated active cases through time. The plot's red line shows the estimated active cases in Tapachula (EAC), calculated as a moving 14-day average of daily new cases officially reported. Copy number/mL quantified in four sites: COAPATAP-WTP (PAE1, black circles), Cahoacán river upstream Tapachula (R1, blue squares), Texcuyuapan river—inside the city (R2, orange triangles), and sewage near COVID-19 Hospital (H1, green diamonds)
According to the map in Fig. 1, we can appreciate that river samples within the city (R2) and the WTP influent (i.e., PA1E) showed the highest copy number for N1. Interestingly, water samples from rivers outside the city (R1, R5, R6) also had a low N1 copy number. In contrast, the sample R3 collected near the city’s WTP contained an intermediate N1 copy number with a 41.6% positivity (Table 1).
Correlation Analysis
We calculated non-parametric correlations between the physicochemical parameters, N1 and N2 copy number (copy number/mL), hospital occupancy, and confirmed cases official registry. Our results suggest that in the H1 site, there is a positive correlation (0.91) between the copy number/mL and bed occupancy (e.g., the higher the copy number/mL with the higher bed occupancy percentage), as well as the number of confirmed cases and pH, DO, TOC, TC in samples collected from sewage near the COVID-19 hospital (Fig. 4). N1 and N2 also correlated between the waste treatment plant site (PA1E) and R2 (Figs. 5 and 6, respectively). Marker N2 showed correlations with bed occupancy in the state and the number of confirmed cases in all sites except R1 (Supplementary Figs. S1–S9).
Spearman correlations of physicochemical parameters, COVID-19 epidemic indicators and quantified SARS-CoV-2 gene copy number for N1 and N2 markers in wastewater from the COVID-19 hospital Tapachula, Mexico (site H1). Positive correlations are displayed in blue and negative correlations in red color. The color intensity and the size of the circle are proportional to the correlation coefficients. Temp temperature; EC electric conductivity; DO dissolved oxygen, TOC total organic carbon; TC total carbon; IC inorganic carbon; Copies_n1 N1 SARS-CoV-2 copy number/mL; Copies_n2 N2 SARS-CoV-2 copy number/mL; %City bed occupancy in Tapachula; %State bed occupancy in Chiapas; Positives officially reported new case on the date of sampling
Spearman correlation of physicochemical parameters, COVID-19 epidemic indicators and quantified SARS-CoV-2 copy number/mL for N1 and N2 markers in wastewater from the Texcuyuapan river branch (site R2). Positive correlations are displayed in blue and negative correlations in red color. The color intensity and the size of the circle are proportional to the correlation coefficients. Temp temperature; EC: electric conductivity; DO dissolved oxygen; TOC total organic carbon; TC total carbon; IC inorganic carbon; Copies_n1 N1 SARS-CoV-2 copy number/mL; Copies_n2 N2 SARS-CoV-2 copy number/mL; %City bed occupancy in Tapachula; %State bed occupancy in Chiapas; Positives officially reported new case on the date of sampling
Spearman correlation of physicochemical parameters, COVID-19 epidemic indicators, and quantified SARS-CoV-2 gene copy number for N1 and N2 markers in wastewater from the wastewater treatment plant (WTP) of Tapachula WTP-COAPATAP (site PA1E). Positive correlations are displayed in blue and negative correlations in red color. The color intensity and the size of the circle are proportional to the correlation coefficients. Temp temperature; EC: electric conductivity; DO dissolved oxygen; TOC total organic carbon; TC total carbon; IC inorganic carbon; Copies_n1 N1 SARS-CoV-2 copy number/mL; Copies_n2 N2 SARS-CoV-2 copy number/mL; %City bed occupancy in Tapachula; %State bed occupancy in Chiapas; Positives officially reported new case on the date of sampling
Another interesting finding is the possible negative correlation between the copy number/mL of N1 and N2 with most of the sampled sites’ water temperature (i.e., the higher temperature, the fewer copy number/mL of N1 and N2). We found a positive correlation for pH and copy number/mL in both markers for H1, PA1E, and R2, but only for N2 in sites R4, R5, and R6 (Figs. 4, 5, 6 and Supplementary Figs. S4, S5, S6). There seems to be a positive correlation between dissolved oxygen (DO) and the number of copies of both markers, mainly with N2, in sites R1, R2, R3, R6, and H1 (Figs. 4, 5 and Supplementary Figs. S1, S3, and S6), something similar happens with EC. It is striking that only two sites showed a significant negative correlation (R4 and R5) in DO and copy number/mL (Supplementary Figs. S4 and S5).
Discussion
Several studies have demonstrated the presence of SARS-CoV-2 in wastewater around the world, usually in samples collected in sewage or WTPs. This virus in rivers has only been documented in Ecuador and in remote areas in Brazil, where sanitation services are not widely available (Fongaro et al., 2021; Guerrero-Latorre et al., 2020). A similar scenario exists in Mexico and other tropical areas in low- and medium-income countries. We detected SARS-CoV-2 RNA in all the water samples analyzed (wastewater and rivers). We found a higher copy number/mL of SARS-CoV-2 in water samples collected from rivers near human settlements and in WTPs than samples collected from rivers outside the city, possibly due to human contamination (Table 1).
Our data followed the trend of active case numbers reported by the Ministry of Health in Mexico. The trend is more evident in PAE1, R2, and H1 samples corresponding to the COAPATAP-WTP, an urban river, and sewage outside the hospital. A positive correlation was detected between copy number/mL and SARS-CoV-2 markers (N1 and N2). Similarly, both markers correlated to the number of confirmed cases on the sampling date, probably due to the closeness of sample sites to sewage discharge. However, these trends are not equally evident in all sampling sites, maybe due to each site’s dynamics of moving water (river dynamics). Other WBE studies have reported variations among local trends (Melvin et al., 2021).
We observed that the SARS-CoV-2 RNA copy number in PAE1 increased earlier than the reported number of cases, whereas, in R2, it increased slightly after the peak in the number of reported cases. The presence of non-reported asymptomatic carriers, SARS-CoV-2 RNA degradation during transport in the wastewater system, low availability of clinical testing, and delays in reporting could contribute to these observed differences among sites and concerning the reported number of cases (Bibby et al., 2021).
We detected a lower copy number for N1 and N2 markers compared to that reported in Quito, Ecuador (N1: 2.84 E+2 to 3.19 E+3 copy number/mL and N2: 2.07 E+2 to 2.23 E+3 copy number/mL; Guerrero-Latorre et al., 2020), a remote zone in Brazil (1.1 E+2 and 1.7 E+3 copy number/mL, N1 and N2 average; Fongaro et al., 2021) or Monterrey Metropolitan Area in the north of Mexico (N1: 2.5 to 7.0 copy number/mL; Mahlknecht et al., 2021). On the other hand, we detected similar or higher copy numbers than those seen in rivers and irrigation channels in central Mexico (8.0–1.97 E+2 copy number/mL; Coronado et al., 2021; González-Reyes et al., 2021) or wastewater in Quintana Roo, Mexico (2.4 E−1–7.5 copy number/mL; Rosiles-González et al., 2021). Our method proved suitable for low copy number samples (N1 had a range of detection from 3.0 to 6.57 E+2 copy number/mL, and N2 was from 6.0 to 7.38 E+2 copy number/mL). Variations in results from other studies probably reflect differences in population density in several cities (Quito 3 million inhabitants, Monterrey 5.3 million, and Tapachula 350,000 inhabitants). Mexico City and the metropolitan area have more than 10 million inhabitants. However, we detected similar gene copy numbers in the Tapachula rivers. These differences could be explained by the distance between the SARS-CoV-2 source and sampling site (Coronado et al., 2021), local epidemic, population dynamics at sampling time, and wastewater treatment infrastructure.
A border city like Tapachula has a changing population due to different migratory movements (Eche, 2013). Indeed, around one million people, residing or in transit, were in Tapachula in 2020, 300% more than the reported census data (COMAR, 2021; INEGI, 2020). The Ministry of Health recorded 1,371 patients infected (recovered) and 252 deaths for the region in of 2020, probably underestimating the number of cases. Tracking COVID-19 cases at the individual level in the area is challenging. On the one hand, the migrant population usually finds it difficult to access proper health and sanitation facilities. Additionally, at the time of our study, only a couple of institutions in Tapachula were qualified to report COVID-19 cases to the Ministry of Health to be included in the official count. However, everyone (symptomatic and asymptomatic carriers) potentially contributes to the viral load in sewage and urban rivers.
Similarly, rivers receive wastewater from rural communities that lack wastewater treatment infrastructure, which is a significant concern in Chiapas, and southern Mexico as most of the population inhabits rural areas. The same phenomenon is observed in other low- and medium-income countries (de Oliveira et al., 2021), where health care infrastructure is insufficient for the population and often only available in cities. Therefore, monitoring rivers for SARS-CoV-2 RNA with an inexpensive method should be informative enough when Population-Based Epidemiology is not optimal.
Our results show that even at the site upstream from the city (R1), in an area with a considerably lower population density, it was possible to detect viral loads of 3 to 123 copy number/mL for the N1 marker on some sampling dates. Control PMMoV was not detected in these samples, which might indicate lower fecal contamination in this collection point (Fig. 2) at that specific sampling period. PCR inhibitors in these samples were ruled out. These findings reveal that rivers are dynamic systems with variations in virus presence through different seasons and still can reflect SARS-CoV-2 prevalence in some instances.
All measurements (pH, temperature, electrical conductivity, salinity, dissolved oxygen, total organic carbon, total carbon, and inorganic carbon) met the established standards (see Table 1). Thus, we suggest that it is unlikely that any parameters could have had an inhibitory effect on PCR reactions. However, we did not measure specific compounds such as residual chlorine or metals in raw wastewater, which may be influencing viral particle integrity or negatively affecting PCR. For example, Greaves et al. (2022) reported that the disinfectant sodium hypochlorite (free chlorine) efficiently inactivates SARS-CoV-2 in water solutions. Moreover, Chin et al. (2020) observed that this virus is stable at room temperature in a wide pH range (3–10). However, they could not find infectious viral particles after a 5 min incubation with hand soap.
Although there was a negative correlation between SARS-CoV-2 RNA copy number/mL and temperature, our results suggest that even in the warm conditions of the tropical environments (water temperature in rivers ranged from 21.3 to 25.8 °C, and COAPATAP-WTP reached up to 29 °C), virus detection and quantification were possible. A similar trend was observed in experiments where T90 (the time to achieve 90% reduction from the initial titter) for SARS-CoV-2 persistence in river water was 1.9 days at 24 °C and 1.2 days in wastewater. In contrast, at 4 °C, T90 was 7.7 and 5.5 days in river water and wastewater, respectively (de Oliveira et al., 2021). Our results on SARS-CoV-2 detection are relevant for other tropical regions where water temperatures are high.
Due to their location in urban sites, we expected similar dynamics from river sites R2 and R4 (sites within Tapachula city); however, the correlation results were different (Fig. 4 and Supplementary Fig. S4) to various contaminants sources occurring along the rivers. More research is needed to infer the physicochemical conditions that allow for reliable detection of SARS-CoV-2 in R2 instead of other urban rivers (i.e., R4). It should be noted that the physicochemical parameters of both sites (R2 and R4) are similar except for EC and DO. The first correlated positively to marker N2 in both rivers, but DO shows a strong negative correlation to marker N2 only in the R4 site (in R2, the correlation is positive). Other studies in water from a biological activated sludge tank from WTP found that detections of N1, N2, and N3 genes decreased with DO decrements (Hong et al., 2021). Therefore, DO and EC should be noticed as physicochemical criteria for setting the method’s reliability threshold. Further research and increasing sample number are needed to explore the possible effects of physicochemical parameters on SARS-CoV-2 RNA stability.
Our study applied a suitable method for the concentration and detection of SARS-CoV-2 in water from different sources. Even though viral concentration methods have been in use for 60 years (Albertsson & Frick, 1960), the first coronavirus epidemic in 2004 increased the relevance of these methods with an enveloped virus (Wang et al., 2005). Then and now WBE proved to be an efficient monitoring system that correlates with SARS-CoV-2-confirmed cases and bed occupancy in hospitals. Our results support this finding showing a similar tendency throughout our study, even from urban rivers samples. It would be helpful to sample areas upstream of urban rivers or WTPs to identify local disease outbreaks and transmission as the copy number detected would reflect the number of cases in those communities (Ahmed et al., 2022). We recommend sampling in discharging sites from sewage for urban rivers to avoid RNA dilution in WTP influent.
Further research is needed to achieve better standardization of the methods since we are sampling in dynamic systems (i.e., rivers) in a region with a heavy rain season, which may affect physicochemical properties that affect RNA stability and, thus, detection. Future work should incorporate normalizing and correction strategies to increase the reliability of WBE methods (Bibby et al., 2021; Melvin et al., 2021). An example would be the variability in SARS-CoV-2 RNA shedding by population normalization with PMMoV quantitation as this is a more persistent and stable virus than a human enteric virus (Ahmed et al., 2022; Kitajima et al., 2018). Additionally, when RNA quantity is sufficient, it should be helpful to have six or more replicates per sample to compensate for subsampling error, especially in low copy number/mL samples (Ahmed et al., 2022). In our case, RNA yield was limited due to the sample volume.
Two years into the pandemic, the world suffered from the Omicron variant wave. Strategies for SARS-CoV-2 monitoring are still needed in several low-income countries. Following international guidelines to adapt and improve SARS-CoV-2 and other pathogens detection may lead us to an active wastewater surveillance system (Borchardt et al., 2021).
Conclusion
Our results suggest that SARS-CoV-2 RNA surveillance of wastewater and rivers can serve as a monitoring system for specific tropical areas or an early warning for a possible outbreak. We also show that data generated on SARS-CoV-2 infection in a population with the migratory flow can be monitored by wastewater and water from river samples in tropical regions with limited infrastructure and with inhabitants hard to screen as the floating population.
References
Aguado-García, Y., Taboada, B., Morán, P., Rivera-Gutiérrez, X., Serrano-Vázquez, A., Iša, P., Rojas-Velázquez, L., Pérez-Juárez, H., López, S., Torres, J., Ximénez, C., & Arias, C. F. (2020). Tobamoviruses can be frequently present in the oropharynx and gut of infants during their first year of life. Scientific Reports, 10(1), 13595. https://doi.org/10.1038/s41598-020-70684-w
Ahmed, W., Angel, N., Edson, J., Bibby, K., Bivins, A., O’Brien, J. W., Choi, P. M., Kitajima, M., Simpson, S. L., Li, J., Tscharke, B., Verhagen, R., Smith, W. J. M., Zaugg, J., Dierens, L., Hugenholtz, P., Thomas, K. V., & Mueller, J. F. (2020a). First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Science of the Total Environment, 728, 138764. https://doi.org/10.1016/J.SCITOTENV.2020.138764
Ahmed, W., Bertsch, P. M., Bibby, K., Haramoto, E., Hewitt, J., Huygens, F., Gyawali, P., Korajkic, A., Riddell, S., Sherchan, S. P., Simpson, S. L., Sirikanchana, K., Symonds, E. M., Verhagen, R., Vasan, S. S., Kitajima, M., & Bivins, A. (2020b). Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environmental Research, 191, 110092. https://doi.org/10.1016/j.envres.2020.110092
Ahmed, W., Simpson, S. L., Bertsch, P. M., Bibby, K., Bivins, A., Blackall, L. L., Bofill-Mas, S., Bosch, A., Brandão, J., Choi, P. M., Ciesielski, M., Donner, E., D’Souza, N., Farnleitner, A. H., Gerrity, D., Gonzalez, R., Griffith, J. F., Gyawali, P., Haas, C. N.,…Shanks, O. C. (2022). Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance. Science of the Total Environment, 805, 149877. https://doi.org/10.1016/j.scitotenv.2021.149877
Albertsson, P., & Frick, G. (1960). Partition of virus particles in a liquid two-phase system. Biochimica Et Biophysica Acta, 37(2), 230–237. https://doi.org/10.1016/0006-3002(60)90228-6
Aslan, A., Shah, G., Sittaramane, V., & Shankar, P. (2020). Sewage monitoring in rural communities: A powerful strategy for COVID-19 surveillance. Journal of Environmental Health, 83(5), 8–10.
Berchenko, Y., Manor, Y., Freedman, L. S., Kaliner, E., Grotto, I., Mendelson, E., & Huppert, A. (2017). Estimation of polio infection prevalence from environmental surveillance data. Science Translational Medicine. https://doi.org/10.1126/scitranslmed.aaf6786
Bibby, K., Bivins, A., Wu, Z., & North, D. (2021). Making waves: Plausible lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Research, 202, 117438. https://doi.org/10.1016/j.watres.2021.117438
Bivins, A., Greaves, J., Fischer, R., Yinda, K. C., Ahmed, W., Kitajima, M., Munster, V. J., & Bibby, K. (2020). Persistence of SARS-CoV-2 in water and wastewater. Environmental Science & Technology Letters, 7(12), 937–942. https://doi.org/10.1021/acs.estlett.0c00730
Borchardt, M. A., Boehm, A. B., Salit, M., Spencer, S. K., Wigginton, K. R., & Noble, R. T. (2021). The environmental microbiology minimum information (EMMI) guidelines: QPCR and dPCR quality and reporting for environmental microbiology. Environmental Science and Technology, 55(15), 10210–10223. https://doi.org/10.1021/ACS.EST.1C01767/SUPPL_FILE/ES1C01767_SI_001.PDF
CDC. (2020). CDC 2019-nCoV real-time RT-PCR diagnostic panel (CDC—manufacturer instructions/package insert FDA. Retrieved February 26, 2021, from https://www.fda.gov/media/134922/
Chin, A. W. H., Chu, J. T. S., Perera, M. R. A., Hui, K. P. Y., Yen, H.-L., Chan, M. C. W., Peiris, M., & Poon, L. L. M. (2020). Stability of SARS-CoV-2 in different environmental conditions. The Lancet Microbe, 1(1), e10. https://doi.org/10.1016/S2666-5247(20)30003-3
COMAR. (2021, November 15). La COMAR en números. Gobierno de Mexico. La COMAR en números. Retrieved November 15, 2021, from https://www.gob.mx/comar/articulos/la-comar-en-numeros-284391?idiom=es
CONAGUA. (2018). Estadísticas del agua en México (2018th ed.). Comisión Nacional del Agua. Retrieved November 6, 2021, from https://sina.conagua.gob.mx/publicaciones/EAM_2018.pdf
CONAGUA. (2020). Inventario de Registros por Década Año, Estaciones climatológicas. Comisión Nacional del Agua. Retrieved February 26, 2021, from https://smn.conagua.gob.mx/tools/RESOURCES/Estadistica/7187.pdf
Coronado, Y., Navarro, R., Mosqueda, C., Valenzuela, V., Pérez, J. P., González-Mendoza, V., de la Torre, M., & Rocha, J. (2021). SARS-CoV-2 in wastewater from Mexico City used for irrigation in the Mezquital Valley: Quantification and modeling of geographic dispersion. Environmental Management, 68(4), 1. https://doi.org/10.1007/S00267-021-01516-4
Davó, L., Seguí, R., Botija, P., Beltrán, M. J., Albert, E., Torres, I., López-Fernández, P. Á., Ortí, R., Maestre, J. F., Sánchez, G., & Navarro, D. (2021). Early detection of SARS-CoV-2 infection cases or outbreaks at nursing homes by targeted wastewater tracking. Clinical Microbiology and Infection. https://doi.org/10.1016/j.cmi.2021.02.003
de Oliveira, L. C., Torres-Franco, A. F., Lopes, B. C., Santos, B. S. Á. S., Costa, E. A., Costa, M. S., Reis, M. T. P., Melo, M. C., Polizzi, R. B., Teixeira, M. M., & Mota, C. R. (2021). Viability of SARS-CoV-2 in river water and wastewater at different temperatures and solids content. Water Research, 195, 117002. https://doi.org/10.1016/j.watres.2021.117002
Eche, D. (2013). Land degradation, small-scale farms' development, and migratory flows in Chiapas case study: Tapachula. In International Rural Development 4 (Vol. 4). kassel university press GmbH. Retrieved February 8, 2022, from https://books.google.com/books/about/Land_Degradation_Small_Scale_Farms_Devel.html?id=lpTDAgAAQBAJ
Farkas, K., Cooper, D. M., McDonald, J. E., Malham, S. K., de Rougemont, A., & Jones, D. L. (2018). Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Science of the Total Environment, 634, 1174–1183. https://doi.org/10.1016/j.scitotenv.2018.04.038
Fongaro, G., Rogovski, P., Savi, B. P., Cadamuro, R. D., Pereira, J. V. F., Anna, I. H. S., Rodrigues, I. H., Souza, D. S. M., Saravia, E. G. T., Rodríguez-Lázaro, D., & da Silva Lanna, M. C. (2021). SARS-CoV-2 in human sewage and river water from a remote and vulnerable area as a surveillance tool in Brazil. Food and Environmental Virology. https://doi.org/10.1007/s12560-021-09487-9
González-Reyes, J. R., Hernández-Flores, M. L., Paredes-Zarco, J. E., Téllez-Jurado, A., Fayad-Meneses, O., & Carranza-Ramírez, L. (2021). Detection of SARS-CoV-2 in wastewater northeast of Mexico City: Strategy for monitoring and prevalence of COVID-19. International Journal of Environmental Research and Public Health, 18(16), 8547. https://doi.org/10.3390/IJERPH18168547
Greaves, J., Fischer, R. J., Shaffer, M., Bivins, A., Holbrook, M. G., Munster, V. J., & Bibby, K. (2022). Sodium hypochlorite disinfection of SARS-CoV-2 spiked in water and municipal wastewater. Science of the Total Environment, 807, 150766. https://doi.org/10.1016/j.scitotenv.2021.150766
Gu, J., Han, B., & Wang, J. (2020). COVID-19: Gastrointestinal manifestations and potential fecal–oral transmission. Gastroenterology, 158(6), 1518–1519. https://doi.org/10.1053/j.gastro.2020.02.054
Guerrero-Latorre, L., Ballesteros, I., Villacrés-Granda, I., Granda, M. G., Freire-Paspuel, B., & Ríos-Touma, B. (2020). SARS-CoV-2 in river water: Implications in low sanitation countries. Science of the Total Environment, 743, 140832. https://doi.org/10.1016/j.scitotenv.2020.140832
Gulino, K., Rahman, J., Badri, M., Morton, J., Bonneau, R., & Ghedin, E. (2020). Initial mapping of the New York City wastewater virome. Msystems. https://doi.org/10.1128/mSystems.00876-19
Hong, P. Y., Rachmadi, A. T., Mantilla-Calderon, D., Alkahtani, M., Bashawri, Y. M., al Qarni, H., O’Reilly, K. M., & Zhou, J. (2021). Estimating the minimum number of SARS-CoV-2 infected cases needed to detect viral RNA in wastewater: To what extent of the outbreak can surveillance of wastewater tell us? Environmental Research, 195, 110748. https://doi.org/10.1016/J.ENVRES.2021.110748
INEGI. (2016). Estudio de informaciòn integrada de la cuenca Rio Suchiate y otras. Retrieved February 26, 2021, from https://www.inegi.org.mx/contenido/productos/prod_serv/contenidos/espanol/bvinegi/productos/nueva_estruc/702825086930_1.pdf
INEGI. (2020). Censo Población y Vivienda 2020. Retrieved February 22, 2022, from https://www.inegi.org.mx/programas/ccpv/2020/#Datos_abiertos
Jørgensen, A. C. U., Gamst, J., Hansen, L. V., Knudsen, I. I. H., & Jensen, S. K. S. (2020). Eurofins Covid-19 SentinelTM wastewater test provide early warning of a potential COVID-19 outbreak. MedRxiv. https://doi.org/10.1101/2020.07.10.20150573
Kitajima, M., Ahmed, W., Bibby, K., Carducci, A., Gerba, C. P., Hamilton, K. A., Haramoto, E., & Rose, J. B. (2020). SARS-CoV-2 in wastewater: State of the knowledge and research needs. Science of the Total Environment, 739, 139076. https://doi.org/10.1016/j.scitotenv.2020.139076
Kitajima, M., Sassi, H. P., & Torrey, J. R. (2018). Pepper mild mottle virus as a water quality indicator. Npj Clean Water, 1(1), 1–9. https://doi.org/10.1038/s41545-018-0019-5
Kocamemi, B. A., Kurt, H., Sait, A., Sarac, F., Saatci, A. M., & Pakdemirli, B. (2020). SARS-CoV-2 detection in Istanbul wastewater treatment plant sludges. MedRxiv. https://doi.org/10.1101/2020.05.12.20099358
Kumar, M., Patel, A. K., Shah, A. V., Raval, J., Rajpara, N., Joshi, M., & Joshi, C. G. (2020). First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Science of the Total Environment, 746, 141326. https://doi.org/10.1016/j.scitotenv.2020.141326
Lago, P. M., Gary, H. E., Pérez, L. S., Cáceres, V., Olivera, J. B., Puentes, R. P., Corredor, M. B., Jímenez, P., Pallansch, M. A., & Cruz, R. G. (2003). Poliovirus detection in wastewater and stools following an immunization campaign in Havana, Cuba. International Journal of Epidemiology, 32(5), 772–777. https://doi.org/10.1093/ije/dyg185
Lescure, F. X., Bouadma, L., Nguyen, D., Parisey, M., Wicky, P. H., Behillil, S., Gaymard, A., Bouscambert-Duchamp, M., Donati, F., le Hingrat, Q., Enouf, V., Houhou-Fidouh, N., Valette, M., Mailles, A., Lucet, J. C., Mentre, F., Duval, X., Descamps, D., Malvy, D.,…Yazdanpanah, Y. (2020). Clinical and virological data of the first cases of COVID-19 in Europe: A case series. The Lancet Infectious Diseases, 20(6), 697–706. https://doi.org/10.1016/S1473-3099(20)30200-0
Mahlknecht, J., Padilla Reyes, D. A., Ramos, E., Reyes, L. M., & Álvarez, M. M. (2021). The presence of SARS-CoV-2 RNA in different freshwater environments in urban settings determined by RT-qPCR: Implications for water safety. Science of the Total Environment, 784, 147183. https://doi.org/10.1016/j.scitotenv.2021.147183
Medema, G., Heijnen, L., Elsinga, G., Italiaander, R., & Brouwer, A. (2020). Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environmental Science & Technology Letters, 7(7), 511–516. https://doi.org/10.1021/acs.estlett.0c00357
Melvin, R. G., Hendrickson, E. N., Chaudhry, N., Georgewill, O., Freese, R., Schacker, T. W., & Simmons, G. E. (2021). A novel wastewater-based epidemiology indexing method predicts SARS-CoV-2 disease prevalence across treatment facilities in metropolitan and regional populations. Scientific Reports, 11(1), 21368. https://doi.org/10.1038/s41598-021-00853-y
Michael-Kordatou, I., Karaolia, P., & Fatta-Kassinos, D. (2020). Sewage analysis as a tool for the COVID-19 pandemic response and management: The urgent need for optimised protocols for SARS-CoV-2 detection and quantification. Journal of Environmental Chemical Engineering, 8(5), 104306. https://doi.org/10.1016/J.JECE.2020.104306
Nemudryi, A., Nemudraia, A., Wiegand, T., Surya, K., Buyukyoruk, M., Cicha, C., Vanderwood, K. K., Wilkinson, R., & Wiedenheft, B. (2020). Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. Cell Reports Medicine. https://doi.org/10.1016/j.xcrm.2020.100098
Pan, Y., Zhang, D., Yang, P., Poon, L. L. M., & Wang, Q. (2020). Viral load of SARS-CoV-2 in clinical samples. The Lancet Infectious Diseases, 20(4), 411–412. https://doi.org/10.1016/S1473-3099(20)30113-4
Pandey, D., Verma, S., Verma, P., Mahanty, B., Dutta, K., Daverey, A., & Arunachalam, K. (2021). SARS-CoV-2 in wastewater: Challenges for developing countries. International Journal of Hygiene and Environmental Health, 231, 113634. https://doi.org/10.1016/j.ijheh.2020.113634
Peccia, J., Zulli, A., Brackney, D. E., Grubaugh, N. D., Kaplan, E. H., Casanovas-Massana, A., Ko, A. I., Malik, A. A., Wang, D., Wang, M., Warren, J. L., Weinberger, D. M., Arnold, W., & Omer, S. B. (2020). Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nature Biotechnology, 38(10), 1164–1167. https://doi.org/10.1038/s41587-020-0684-z
Philo, S. E., Keim, E. K., Swanstrom, R., Ong, A. Q. W., Burnor, E. A., Kossik, A. L., Harrison, J. C., Demeke, B. A., Zhou, N. A., Beck, N. K., Shirai, J. H., & Meschke, J. S. (2021). A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. The Science of the Total Environment, 760, 144215. https://doi.org/10.1016/j.scitotenv.2020.144215
R Core Team. (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing. Retrieved November 1, 2020, from https://www.r-project.org/
Randazzo, W., Truchado, P., Cuevas-Ferrando, E., Simón, P., Allende, A., & Sánchez, G. (2020). SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Research, 181, 115942. https://doi.org/10.1016/j.watres.2020.115942
Rimoldi, S. G., Stefani, F., Gigantiello, A., Polesello, S., Comandatore, F., Mileto, D., Maresca, M., Longobardi, C., Mancon, A., Romeri, F., Pagani, C., Cappelli, F., Roscioli, C., Moja, L., Gismondo, M. R., & Salerno, F. (2020). Presence and infectivity of SARS-CoV-2 virus in wastewaters and rivers. Science of the Total Environment, 744, 140911. https://doi.org/10.1016/j.scitotenv.2020.140911
Rosiles-González, G., Carrillo-Jovel, V. H., Alzate-Gaviria, L., Betancourt, W. Q., Gerba, C. P., Moreno-Valenzuela, O. A., Tapia-Tussell, R., & Hernández-Zepeda, C. (2021). Environmental surveillance of SARS-CoV-2 RNA in wastewater and groundwater in Quintana Roo, Mexico. Food and Environmental Virology, 13(4), 457–469. https://doi.org/10.1007/s12560-021-09492-y
Secretaría de Comercio y Fomento Industrial, SECOFI. (1980). Norma Mexicana NMX-AA-003-1980, Aguas residuals. Muestreo. Residual waters sampling. Diario Oficial de la Federación, November 6 1992. Retrieved March 7, 2022, from https://www.gob.mx/cms/uploads/attachment/file/166762/NMX-AA-003-1980.pdf
Serrano-Rivero, Y., Hernández-García, A., & Fando-Calzada, R. (2013). Comparación de dos métodos para la preparación de células competentes en Escherichia coli. Revista CENIC: Ciencias Biológicas, 44(2), 1–6.
Song, Y., Liu, P., Shi, X. L., Chu, Y. L., Zhang, J., Xia, J., Gao, X. Z., Qu, T., & Wang, M. Y. (2020). SARS-CoV-2 induced diarrhoea as onset symptom in patient with COVID-19. Gut, 69(6), 1143–1144. https://doi.org/10.1136/gutjnl-2020-320891
Street, R., Malema, S., Mahlangeni, N., & Mathee, A. (2020). Wastewater surveillance for Covid-19: An African perspective. Science of the Total Environment, 743, 140719. https://doi.org/10.1016/j.scitotenv.2020.140719
Wang, X.-W., Li, J.-S., Guo, T.-K., Zhen, B., Kong, Q.-X., Yi, B., Li, Z., Song, N., Jin, M., Wu, X.-M., Xiao, W.-J., Zhu, X.-M., Gu, C.-Q., Yin, J., Wei, W., Yao, W., Liu, C., Li, J.-F., Ou, G.-R.,…Li, J.-W. (2005). Excretion and detection of SARS coronavirus and its nucleic acid from digestive system. World Journal of Gastroenterology, 11(28), 4390–4395. https://doi.org/10.3748/wjg.v11.i28.4390
Westhaus, S., Weber, F.-A., Schiwy, S., Linnemann, V., Brinkmann, M., Widera, M., Greve, C., Janke, A., Hollert, H., Wintgens, T., & Ciesek, S. (2021). Detection of SARS-CoV-2 in raw and treated wastewater in Germany: Suitability for COVID-19 surveillance and potential transmission risks. The Science of the Total Environment, 751, 141750. https://doi.org/10.1016/j.scitotenv.2020.141750
Wölfel, R., Corman, V. M., Guggemos, W., Seilmaier, M., Zange, S., Müller, M. A., Niemeyer, D., Jones, T. C., Vollmar, P., Rothe, C., Hoelscher, M., Bleicker, T., Brünink, S., Schneider, J., Ehmann, R., Zwirglmaier, K., Drosten, C., & Wendtner, C. (2020). Virological assessment of hospitalized patients with COVID-2019. Nature, 581(7809), 465–469. https://doi.org/10.1038/s41586-020-2196-x
Wu, F., Zhang, J., Xiao, A., Gu, X., Lee, W. L., Armas, F., Kauffman, K., Hanage, W., Matus, M., Ghaeli, N., Endo, N., Duvallet, C., Poyet, M., Moniz, K., Washburne, A. D., Erickson, T. B., Chai, P. R., Thompson, J., & Alm, E. J. (2020). SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. Msystems. https://doi.org/10.1128/mSystems.00614-20
Wurtzer, S., Marechal, V., Mouchel, J. M., Maday, Y., Teyssou, R., Richard, E., Almayrac, J. L., & Moulin, L. (2020). Evaluation of lockdown effect on SARS-CoV-2 dynamics through viral genome quantification in waste water, Greater Paris, France, 5 March to 23 April 2020. Eurosurveillance. https://doi.org/10.2807/1560-7917.ES.2020.25.50.2000776
Xagoraraki, I., & O’Brien, E. (2020). Wastewater-based epidemiology for early detection of viral outbreaks. In D. J. O’Bannon (Ed.), Women in water quality: investigations by prominent female engineers (pp. 75–97). Springer International Publishing. https://doi.org/10.1007/978-3-030-17819-2_5
Xiao, F., Tang, M., Zheng, X., Liu, Y., Li, X., & Shan, H. (2020). Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology, 158(6), 1831-1833.e3. https://doi.org/10.1053/j.gastro.2020.02.055
Zhou, J.-B., Kong, W.-H., Wang, S., Long, Y.-B., Dong, L.-H., He, Z.-Y., & Liu, M.-Q. (2021). Detection of SARS-CoV-2 RNA in medical wastewater in Wuhan during the COVID-19 outbreak. Virologica Sinica, 36(5), 1077–1079. https://doi.org/10.1007/s12250-021-00373-z
Acknowledgements
The authors would like to thank COAPATAP for their help and assistance with sampling. This work was supported by Grant 312724 Proyectos de Investigación Científica, Desarrollo Tecnológico e Innovación en Salud ante la Contingencia por COVID-19 (2020-1), CONACYT, Mexico. We would like to thank Jenifer Villafuerte for her contribution (ECOSUR Project Assistant fellowship).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (DOCX 1620 KB)
BLAST Analyses of Sanger sequences obtained from qPCR products, Sanger sequences, and correlation figures
Rights and permissions
About this article
Cite this article
Zarza, E., Diego-García, E., García, L.V. et al. Monitoring SARS-CoV-2 in the Wastewater and Rivers of Tapachula, a Migratory Hub in Southern Mexico. Food Environ Virol 14, 199–211 (2022). https://doi.org/10.1007/s12560-022-09523-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12560-022-09523-2